Description
GigaEvo is an evolutionary framework for automating ML and LLM-oriented tasks.
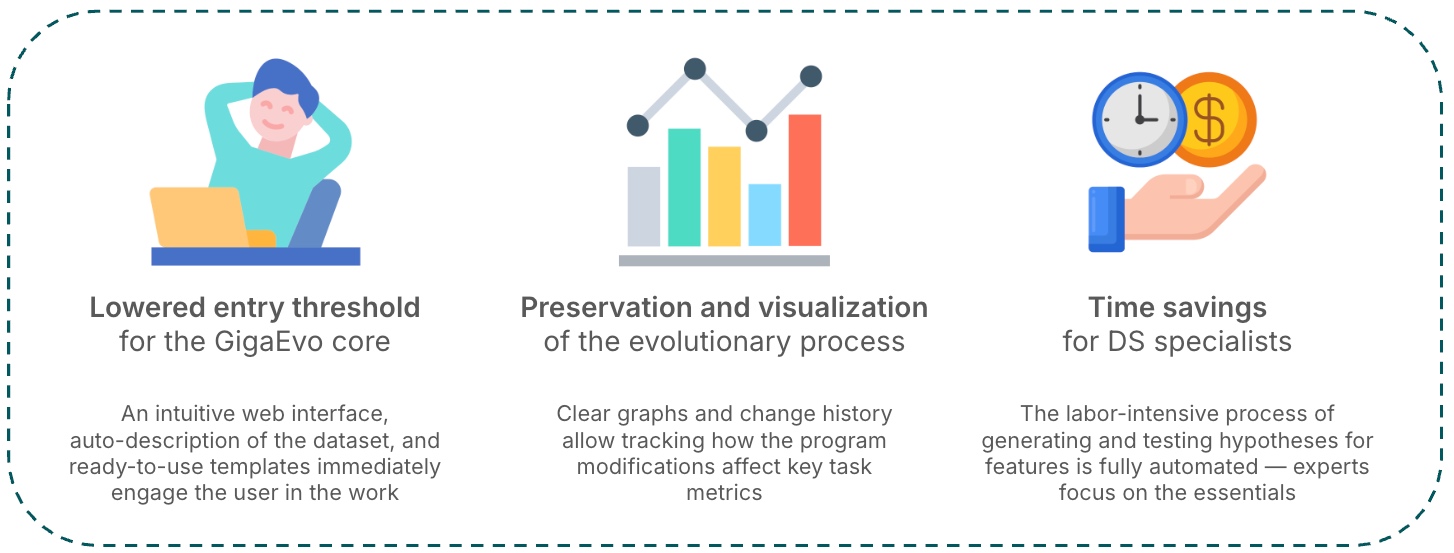
The framework is designed to automate machine learning tasks, including optimization of models, parameters, features, and LLM-oriented methods. The solution minimizes specialist involvement, accelerates the experiment cycle, and enhances the quality of the final models.
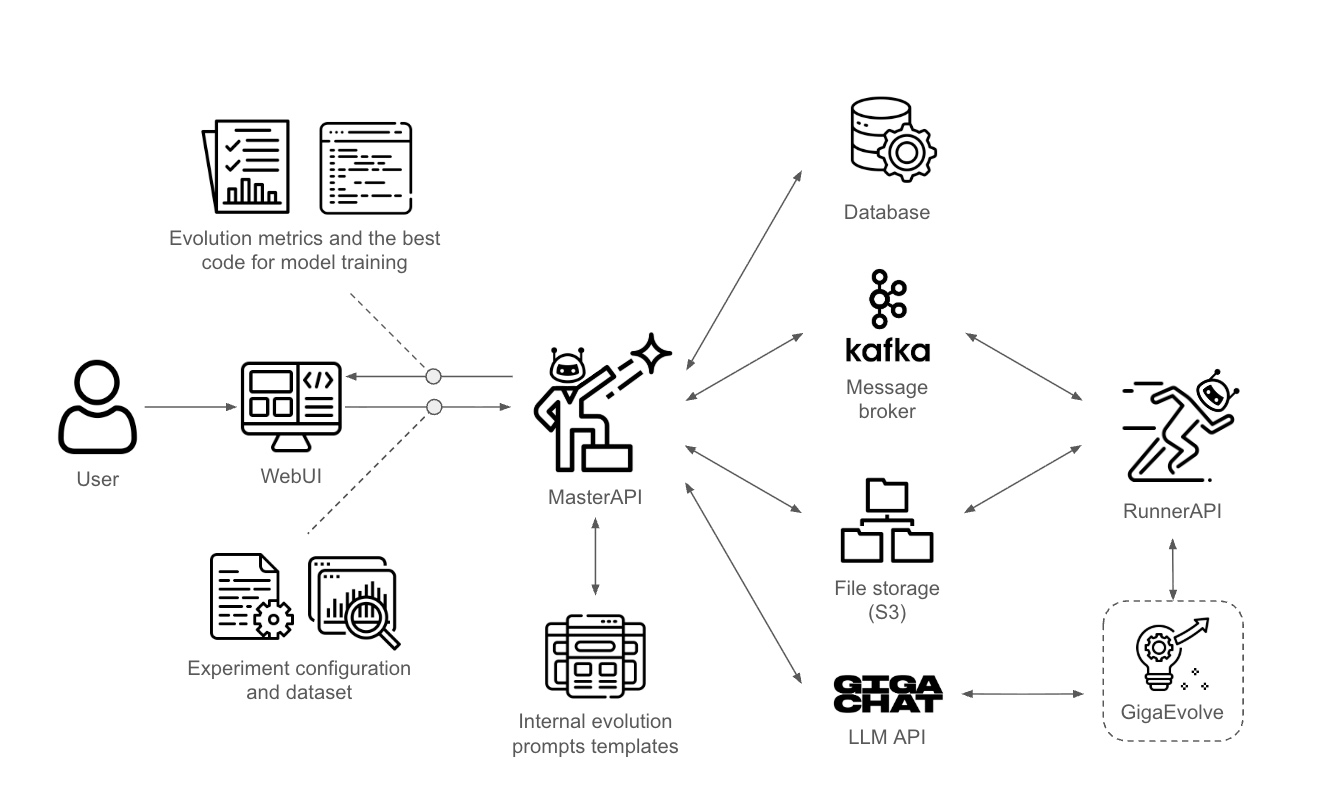
Solution Architecture and Operations
The architecture implements a modular evolutionary approach, where each program is a uniquely identified evolutionary unit containing source code, lifecycle state, metrics, lineage information, and execution results. All data is stored in a Redis layer with support for concurrent access and real-time tracking.
Program execution is organized through an asynchronous framework based on a Directed Acyclic Graph (DAG), implemented in Python using asyncio, which enables parallel execution both between programs and within individual program flows.
Each DAG stage performs independent operations:
- Code execution — running programs
- Validation — verifying result correctness
- Complexity analysis — assessing computational characteristics
- LLM calls — integration with language models
The stages are connected through data transfer and execution order dependencies. The scheduler manages caching and conditional stage skipping.
The asynchronous evolutionary engine implements the MAP-Elites algorithm, maintaining a diverse archive of high-performing programs based on fitness and correctness metrics, including single- and multi-island configurations with periodic solution migration.
The mutation operation is performed by an agent based on LangGraph, which generates prompts using task context, parent programs, and metrics, supporting both diff-based and rewritten mutations, as well as routing between multiple LLMs.
Configuration and experiment management are handled using Hydra, which employs hierarchical YAML files for reproducibility and quick experiment setup.
New optimization tasks are defined through a specification directory containing task descriptions, declarative metric schemas, validation scripts, and source programs, ensuring task definition independence from the computational core and simplifying the integration of new domains.
The system allows multiple experiments to be run simultaneously through the concurrent operation of several RunnerAPI instances, managed by a main module called the Master API. Upon receiving a request, the management service stores all experiment metadata and the dataset to ensure reproducibility of results.
GigaEvo implements a microservice architecture with three main components that work together to provide scalable management of machine learning experiments:
- Master API runs on port 8000 and serves as the central orchestration service responsible for coordinating and managing experiments.
- Multiple Runner API instances run on ports 8001 and above, providing distributed task execution services that launch the actual experiments.
- WebUI runs on port 7860, offering users a web-based interface for experiment creation and real-time progress monitoring.
PostgreSQL serves as the primary database for storing experiment configurations and lifecycle status information. It manages metadata of runner instances, including health status and availability, maintaining a comprehensive registry of all system components. The database retains a complete history of task executions for audit and performance analysis purposes. System audit logs are stored to ensure full traceability of all operations and state changes.
Redis provides high-performance caching and coordination capabilities across the system. It manages task queues and worker coordination, ensuring efficient workload distribution and real-time task status tracking. Real-time status caching provides fast access to experiment and system state information, while session management supports user context and authentication state. Temporary data storage in Redis supports intermediate computation results and transient system state.
MinIO provides S3-compatible object storage for all file data within the system. Input data files and datasets are stored efficiently with metadata for easy retrieval and organization. Experiment results and generated artifacts are saved with comprehensive indexing for future analysis. Generated visualizations and charts are stored in standard formats accessible through the WebUI, while model checkpoints and training outputs support experiment reproducibility and further analysis.
Local File System provides temporary storage for working data during experiment execution. Clones of the GigaEvolve repository are maintained locally for fast access and to support version-controlled experiment environments. Temporary experiment files are created and managed during runtime, providing workspace for intermediate computations. Local computation results are cached to reduce network traffic and improve performance for frequently requested data.
Parallel Execution with Multiple Runners
The Master API automatically deploys and manages Runner API instances as Docker containers, enabling seamless scaling. Each Runner instance operates independently with its own dedicated set of worker processes, ensuring isolation and resource management. New instances can be added dynamically through configuration changes without requiring system restarts, allowing flexible scaling based on demand.
Example runner configuration:
runner_config:
max_workers_per_instance: 5 # Workers per runner instance
auto_initialize: true # Automatic container deployment
instances:
local:
host: "runner-api-1"
is_local: true
remote-1:
host: "remote-server.example.com"
is_local: false
remote-2:
host: "another-server.example.com"
is_local: falseThe Master API tracks real-time status information for all Runner instances, enabling intelligent experiment assignment. Experiments are automatically allocated to available instances with sufficient capacity, ensuring optimal resource utilization. The load-balancing algorithm takes into account the current workload and the health status of each instance when making assignment decisions. Faulty instances are automatically detected and removed from the active rotation, maintaining system reliability.
The system performs continuous health checks every 30 seconds to monitor the status of all Runner instances. Automatic recovery mechanisms are triggered when failures are detected, minimizing downtime and ensuring system availability. Real-time status synchronization between Docker containers and the database is supported, providing accurate and up-to-date information about the system state.
Each Runner API instance can execute multiple experiments simultaneously, providing horizontal scalability. The number of workers per instance is configurable, with a default value of five workers, which can be adjusted based on available resources and workload requirements. Workers independently poll the global task queue, ensuring optimal load distribution across all available system resources.
Monitoring
The system provides comprehensive real-time monitoring capabilities that offer full visibility into system operations and experiment progress. Experiment status monitoring delivers live updates on progress and key metrics, allowing users to track experiment execution in detail. System health monitoring continuously checks the status of containers and services to ensure operational reliability. Performance metric tracking provides insights into resource usage and throughput, helping to optimize overall system performance.
Capabilities
The GigaEvo framework provides a fully automated ML experimentation cycle — from data loading to obtaining the optimal solution — using evolutionary search for learning strategies with LLM-based mutations and rigorous validation. The platform offers convenient real-time progress visualization and significantly reduces manual effort: the user defines the goal and provides data, while the system performs all stages automatically. GigaEvo easily integrates with enterprise infrastructure through the Master API and S3/MinIO, scales via distributed Runner instances, task queues, and isolated workspaces, and also supports exporting results to JSON, delivering the optimal program with metadata and storing artifacts in S3.
- Full-cycle automation: GigaEvo provides automation of the entire ML experimentation cycle — from data loading to obtaining the optimal solution — including automatic evolution of learning strategies, where the system independently discovers and improves the most effective approaches.
- Visualization and control: The platform offers real-time progress visualization and control, minimizing the human factor — the developer defines the task, and the system fully manages the experimentation process.
- Integration and scalability: The solution integrates with existing AutoML and MLOps platforms, supports easy deployment and scaling in cloud and enterprise environments, and ensures seamless connection to organizational infrastructure through the MasterAPI module.
The solution is effectively applied across a wide range of tasks that require fast and high-quality creation, testing, and improvement of machine learning models. In development centers and Data Science labs, it automates the experiment cycle and reduces research time; in analytical and forecasting departments, it accelerates model building and improves prediction accuracy; on hypothesis testing platforms, it enables rapid validation of scientific and business assumptions; and in corporate decision-support systems, it serves as a foundation for intelligent modules that enhance management processes. GigaEvo scales easily and is equally effective for scientific tasks as well as applied solutions in business, industry, and finance.
The API provides a REST interface for integrating GigaEvo with external corporate systems. It allows operations for experiment creation — registering configurations, data, and metadata — as well as starting, stopping, and retrieving current statuses. The API supports viewing experiment lists and histories, along with publishing events to Kafka (configuration retrieval, preparation, launch, stop).
The Runner API is designed for direct management of experiment execution: initialization, starting and stopping processes, retrieving statuses and logs, generating visualizations, delivering the best program (code, fitness, metadata), and working with uploaded artifacts, including scanning S3 prefixes. A full list of available endpoints is provided in docs/api_endpoints.md.
GigaEvoML is especially well suited for machine learning workflows that benefit from parallel execution and scalable experimentation. ML model development teams can use the system for parallel training and evaluation of multiple models at once, dramatically reducing development time. Hyperparameter optimization becomes more efficient through simultaneous experiments across different parameter configurations. A/B testing scenarios benefit from parallel comparison of various model configurations under identical conditions. Research workflows in academia and R&D departments can leverage scalable experiment execution for large-scale ML research projects that would be impractical to run sequentially.
The system serves diverse industries with specific machine learning needs. Financial services organizations use GigaEvoML for risk model development and validation, enabling rapid iteration on complex financial models. Healthcare organizations employ the platform to develop and test medical ML models, supporting the creation of diagnostic and treatment optimization systems. E-commerce companies gain from optimizing recommendation systems through parallel experimentation with various algorithms and parameters. Manufacturing organizations utilize the system for predictive maintenance model development, building systems that anticipate equipment failures and optimize maintenance schedules.
Example Project on GigaEvo
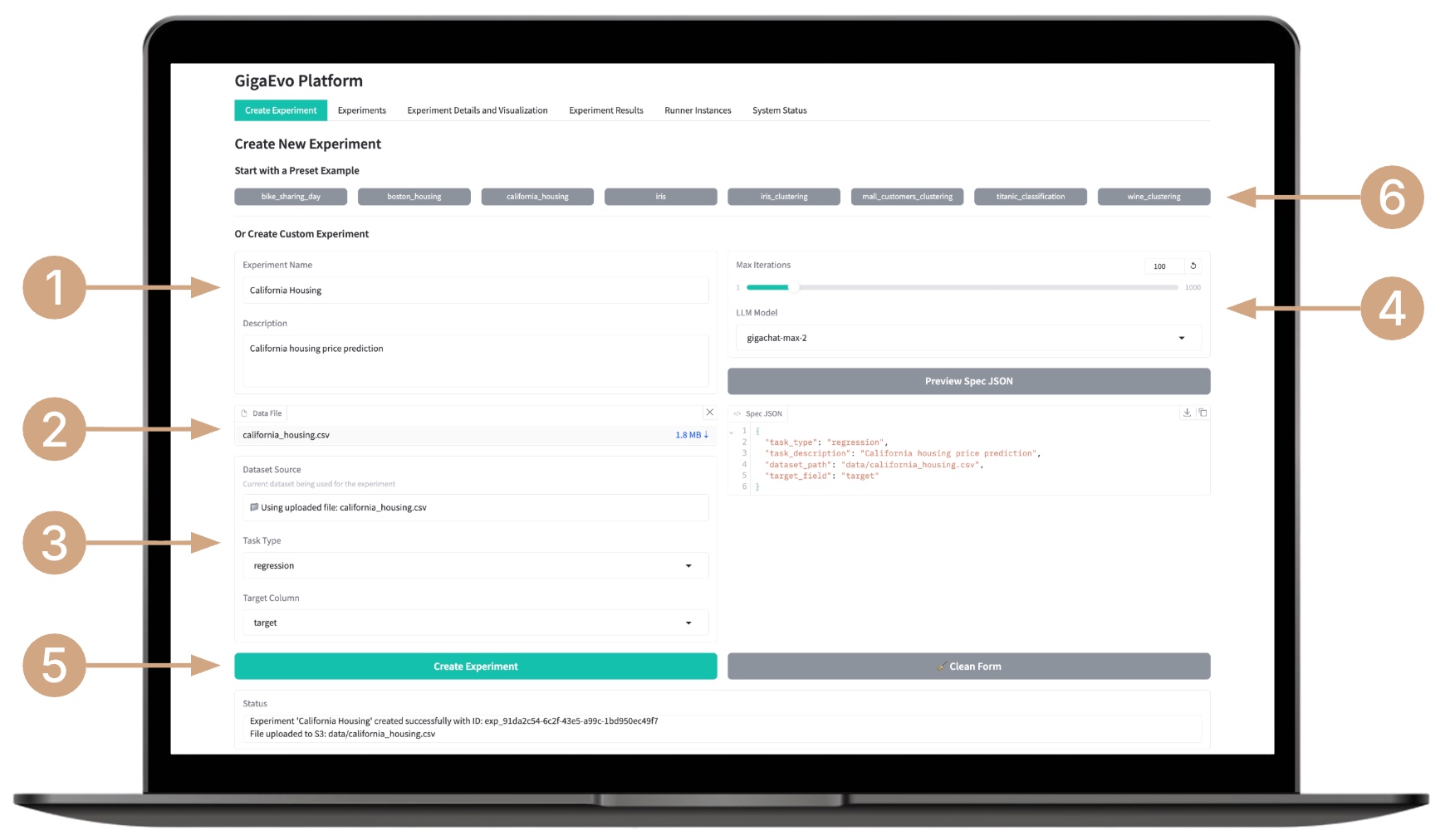
Creating an Experiment
- Enter the experiment name and a brief description of its objective.
- Upload the dataset.
- Select the task type (classification, regression, or clustering), specify target metrics and parameters (for supervised tasks — the target column).
- Configure additional parameters — number of generations, type of LLM model, and other options.
- Create the experiment by clicking the Create Experiment button — the system will automatically assign it a formatted identifier.
- You can choose a template experiment from the list — the system will automatically fill in the necessary fields and upload data; you only need to start it.
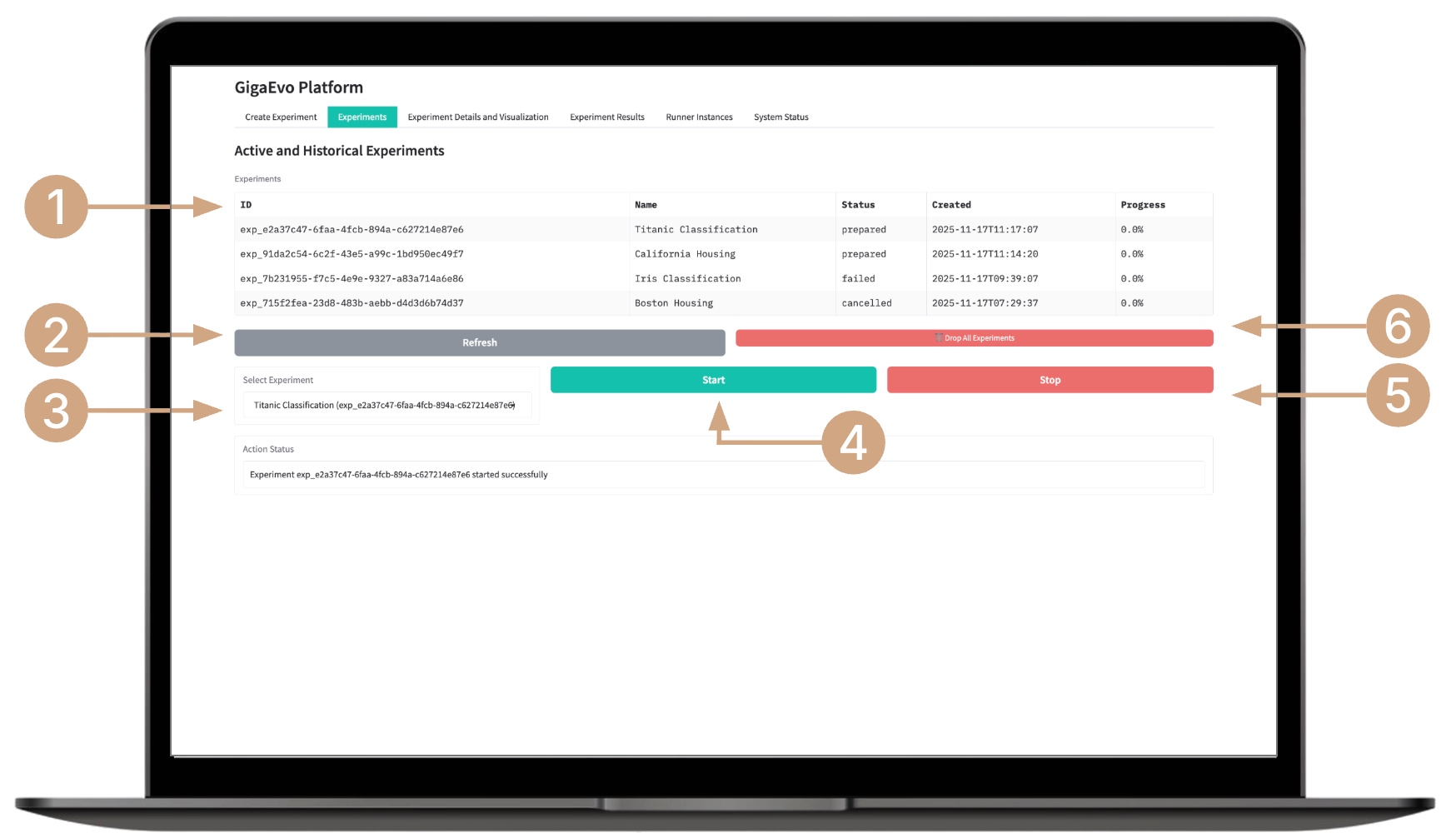
Starting the Experiment
- In the “Experiments” tab, review the list of available experiments and their statuses (PENDING, INITIALIZED, RUNNING, COMPLETED, etc.).
- If necessary, refresh the list by clicking the Refresh button.
- Select an experiment from the list.
- Start the evolution process by clicking the Start button.
- If needed, stop the experiment early by clicking the Stop button.
- You can delete all current experiments by clicking the Drop All Experiments button if you need to clear the list.
Tracking the Experiment
- Select an experiment from the list of available ones — the data will load automatically.
- Monitor key evolution metrics (best metric, best generation, number of iterations and programs) — values update in real time.
- View the evolution dynamics chart — it refreshes automatically.
- If necessary, click the Refresh results button to manually update the data.
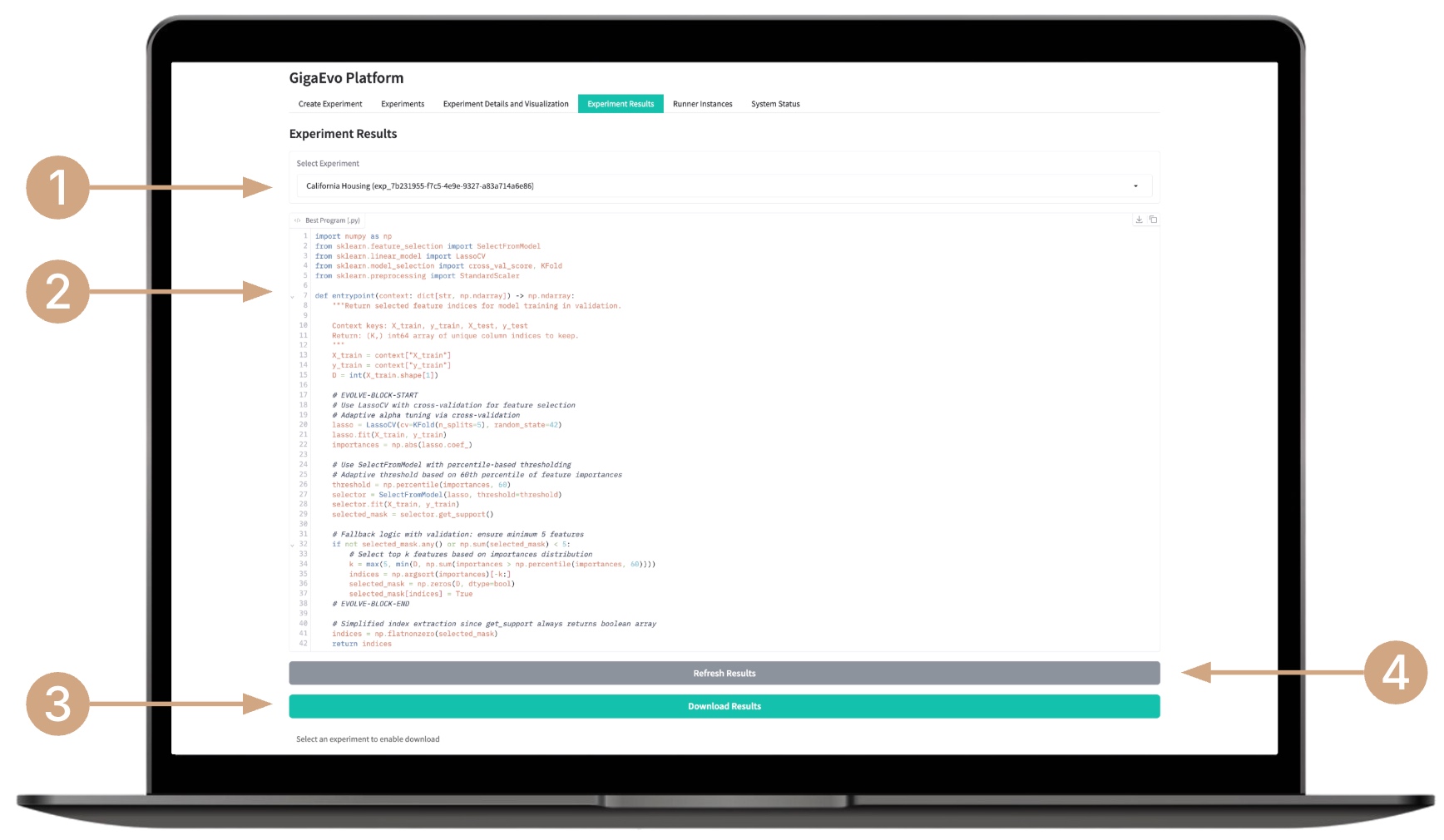
Viewing Experiment Results
- Select an experiment from the list.
- Review the current best program — it is displayed in real time.
- Download the archive containing the best program and the model code used for testing.
- If necessary, click the Refresh results button to manually update the results.
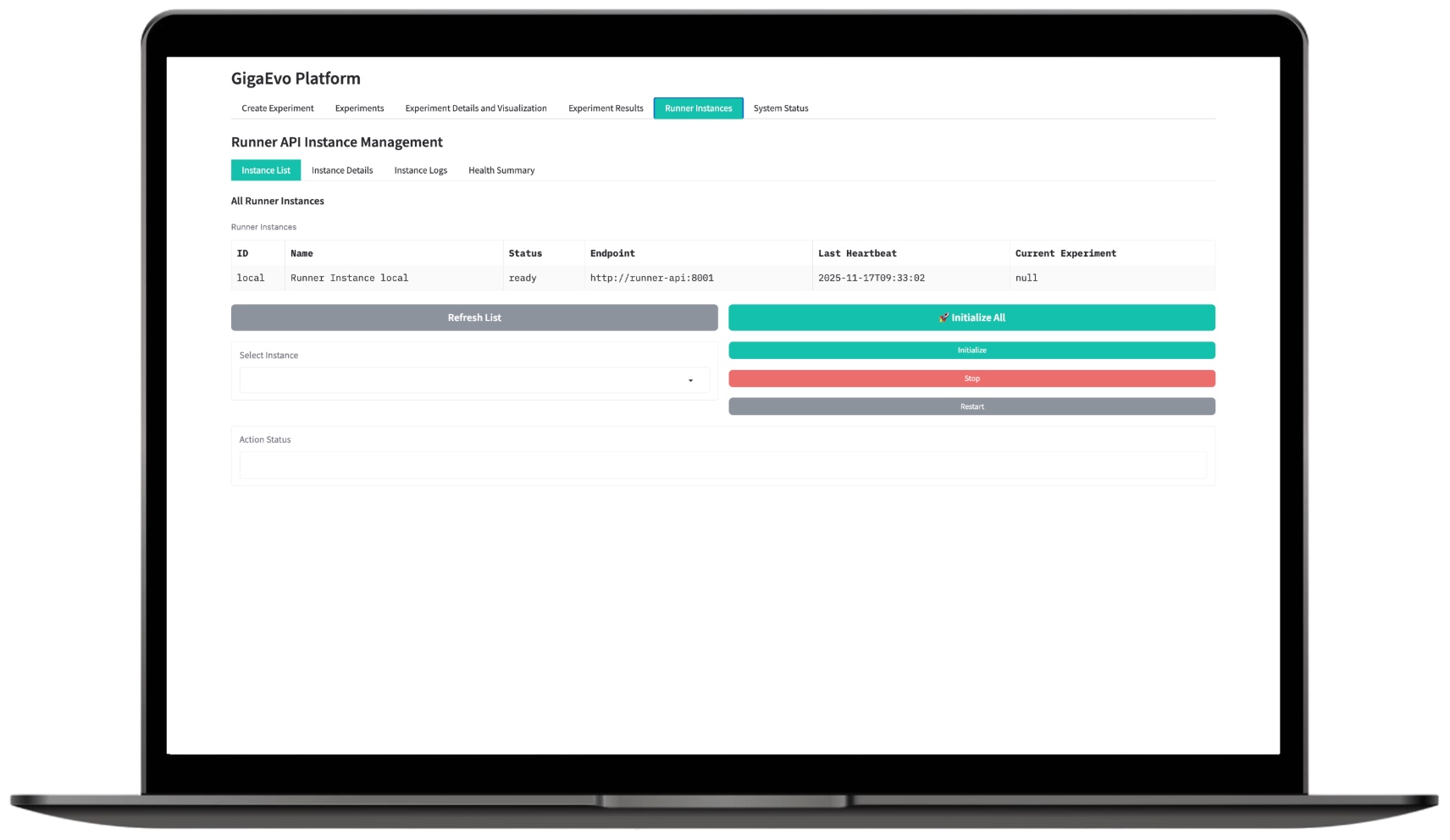
Tracking RunnerAPI Status
This section allows you to monitor the status of all running RunnerAPI workers. The user has access to:
- A list of RunnerAPI instances with their status, endpoint, last heartbeat, and the currently running experiment;
- Basic management operations: Initialize, Stop, Restart;
- Viewing logs of each instance and its health summary;
- Master API endpoints for retrieving the list of instances, details, logs, and overall health summary;
- Direct RunnerAPI health checks (
/health) and experiment status requests on a specific Runner (/api/v1/experiments/{id}/status).
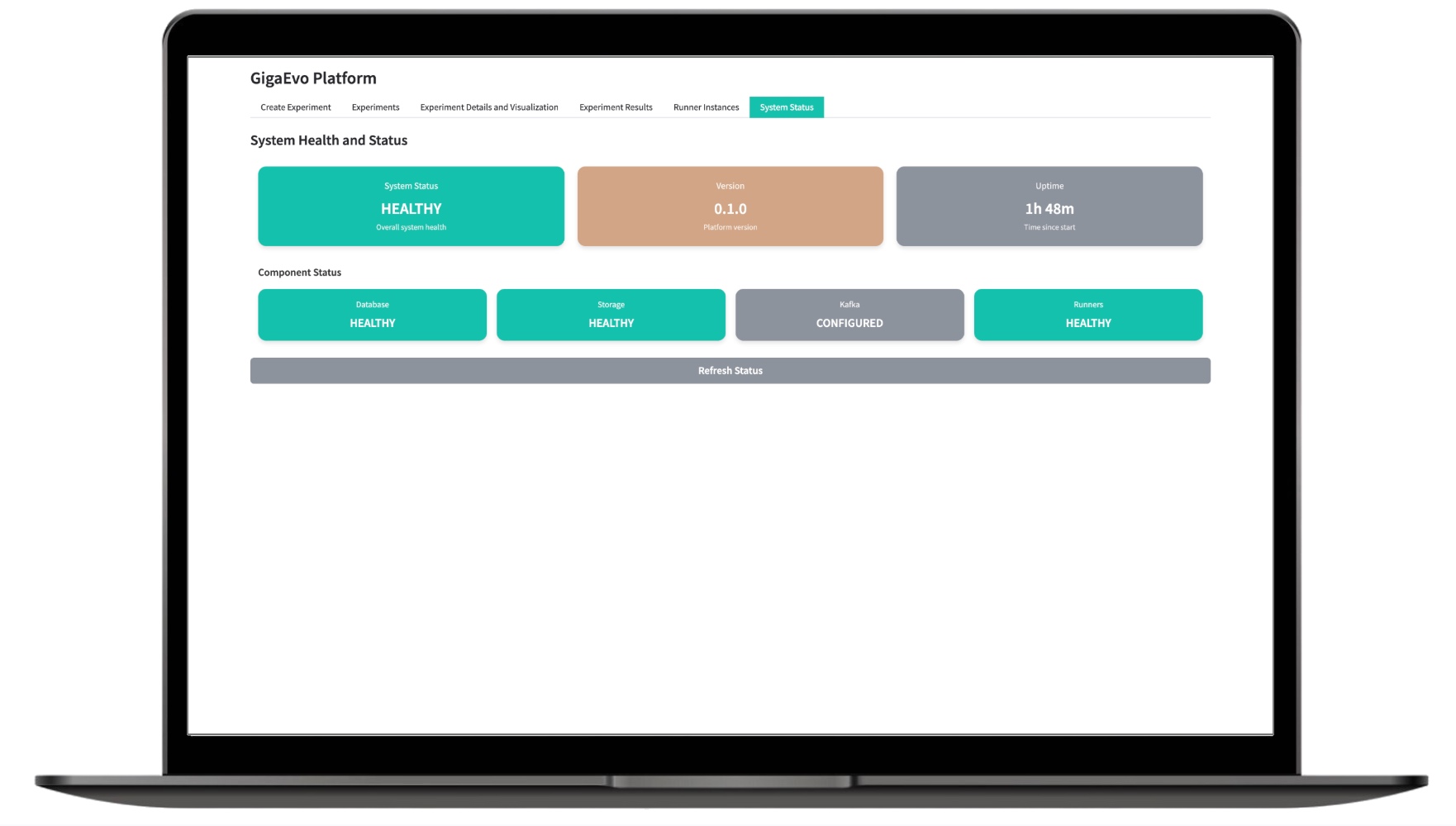
Monitoring system status
The monitoring section displays the current state of all GigaEvo components: from the API to background workers. The user has access to:
- Health check endpoints (
/health) for the master-api, runner-api, and web-ui services; - Logs of background tasks and experiments;
- Status of queues and tasks in Redis;
- Availability checks for the S3 storage and the database.
Using experiment results
- Upload the experiment, start the evolution, and wait to get the best solution based on the metrics.
- Export the experiment results along with the feature engineering code and a model training example.
- Integrate the obtained scripts with the necessary libraries and embed them into your workflows.
Framework Installation
Quick Start
see also README.md
- Requirements: Docker, Docker Compose, Python 3.12+ (for local development),
uvorpip. - LLM models configuration: The platform reads all LLM settings from a single repository-level file:
llm_models.yml. Createllm_models.ymlfrom thellm_models.yml.exampletemplate and fill in your credentials. - To run the whole stack:
make deploy
# или
./deploy.sh deploy- Infrastructure separately:
make deploy-infrastructure- Applications separately:
make deploy-applicationsServices and ports (see docker-compose.yml):
postgres(5432),redis(6379),minio(9000/9001)master-api(8000),runner-api(8001),web-ui(7860)
Health check:
http://localhost:8000/healthhttp://localhost:8001/healthhttp://localhost:7860
1. Installing the GigaEvo Evolution Core
Installing the GigaEvo Evolution Core — installation of the Python package with a full set of dependencies: Redis client, integration with local/remote LLMs, implementation of core pipeline components, analysis tools (pandas/matplotlib).
Necessary dependencies to work with GigaEvo include:
- Core libraries: Redis client, Hydra for configuration management
- LLM integration: OpenAI/OpenRouter API clients, LangChain for LLM operations, prompt formatting utilities
- Evolution components: MAP-Elites implementation, DAG execution engine, automatic program evaluation pipeline
- Analysis tools: Pandas for data export, Matplotlib/Seaborn for visualization
Example execution:
pip install -e .After installation, verify the setup by ensuring that gigaevo can be imported: python -c "import gigaevo; print('OK')". Experiments are run from the root of the repository.
More details: See the "Quick Start" section in README.md
2. Redis Configuration
Redis is a centralized database for the evolution system, storing program states (FRESH → PROCESSING_STARTED → PROCESSING_COMPLETED → EVOLVING), metadata, and execution results. It supports safe coordination between the evolution engine and execution graph via atomic operations and preserves progress for recovery. It contains independent databases — each new run should use a separate DB, specified by the parameter redis.db=N.
Redis serves as the centralized database for the entire evolution system. It stores:
- Program states: All programs pass through states (FRESH → PROCESSING_STARTED → PROCESSING_COMPLETED → EVOLVING)
- Islands archives: Each evolutionary island stores its MAP-Elites grid in Redis with programs indexed by quality and other metadata
- Meta Generation numbers, program quality, additional metrics, execution results
- State indexes: Efficient state search using keys like
state:FRESH:*for programs awaiting evaluation
Multiple databases: Redis supports up to 16 independent databases (indexes 0-15). Each experiment should use its own DB for data isolation. Specify the number using the redis.db=N parameter when launching evolution. This allows parallel experiments without conflicts and preserves results of different runs for later comparison.
Example execution:
# Start the Redis server
redis-server
Redis must run throughout the entire evolution process. If interrupted, evolution can resume from the last saved state since all progress is stored in Redis.
Troubleshooting:
- If the database is not empty: clear the DB with:
redis-cli -n 0 FLUSHDB; or use a different DB:python run.py redis.db=1 - Connection refused: make sure Redis is running on the default port 6379
More details: See the "Quick Start" and "Troubleshooting" sections in README.md for Redis state management
3. Creating an Evolution Task
Creating a task — each evolution task is defined via a directory in problems/<task_name>/. The directory must contain:
task_description.txt: task description for the LLMmetrics.yaml: specification of the target metric and additional metricsvalidate.py: logic for calculating metrics for a new programinitial_programs/: initial programs for evolutionhelper.py(optional): helper functions for programscontext.py(optional): additional input context for programs
Task directory structure: Each task in problems/<task_name>/ requires:
task_description.txt: Description shown to the LLM during mutations (generation of new programs) to understand the task context and goalsmetrics.yaml: Defines the configuration of primary and auxiliary metrics with boundaries, precision, and behavioral space dimensions. Exactly one metric must be primary with the flagis_primary: truevalidate.py: Implements validation logic returning a dictionary with all metrics frommetrics.yamlandis_valid: 1/0(whether the program is valid)initial_programs/: Directory with Python files, each containing anentrypoint()function with an initial program. These form the initial population for evolutionhelper.py(optional): Helper functions for programscontext.py(optional): Additional input context for programs
Creation methods:
- Wizard (recommended): Create the directory using a special wizard (
tools/wizard.py):- Create and fill a YAML config for the task in
tools/config/wizard/(seetools/config/wizard/heilbron.yamlfor example)- The YAML file declaratively describes the task: name, description, list of metrics with boundaries and type, validation parameters, program signature for evolution, and initial programs list.
- The wizard generates all necessary files with TODO comments.
- Run directory creation via
python -m tools.wizard tools/config/wizard/my_config.yaml. The user then implements:- Metric calculation logic in
validate.py - Initial program code in
initial_programs/*.py
- Metric calculation logic in
- Create and fill a YAML config for the task in
- Manually: Create and fill the directory and files yourself. See
problems/heilbron/for a full working example.
Both approaches create identical structures — the wizard automates uniform program creation and ensures consistency.
Creation methods:
Option A: Using the Wizard (recommended)
# Generate from an existing config
python -m tools.wizard heilbron.yaml
# Or create a custom config (see tools/README.md for format)
python -m tools.wizard my_problem.yamlGenerated structure:
problems/my_problem/
├── validate.py # TODO: Implement fitness evaluation
├── metrics.yaml # Metrics specification
├── task_description.txt # Task description
└── initial_programs/
└── strategy1.py # TODO: Implement the entrypoint() functionOption B: Manual creation
mkdir -p problems/my_problem/initial_programs
# Create validate.py, metrics.yaml, task_description.txt, initial_programs/*.py
# Follow the structure described above and use problems/heilbron/ as an exampleMore details: See the "Advanced Usage" section (subsections "Generate Problem with Wizard" and "Create Your Own Problem Manually") in README.md, as well as tools/README.md for the wizard YAML config format
4. Evolution Launch
Launching evolution is an iterative optimization process: loading initial programs → evaluation through a DAG pipeline → selection of the best solutions → mutation using LLM → repetition for multiple generations. The pipeline uses Hydra for modular evolution configuration management with prepared templates. Results are logged to outputs/, while data is stored in the Redis DB.
Evolution launch is done by supplying yaml config files in the config/ directory. The pipeline uses Hydra for modular config management. All settings in config/ can be composed and overridden. Experiment configs (config/experiment/) provide preconfigured templates for evolution.
Available experiments:
base– single island evolution (default)multi_island_complexity– two islands: performance + simplicitymulti_llm_exploration– multiple LLMs for diverse mutationsfull_featured– multi-island + multi-LLM combined
Examples of other parameters to override:
max_generations=N– number of evolution iterationstemperature=0.0-2.0– LLM creativity (higher = more creative)redis.db=N– Redis DB number (useful for parallel runs)model_name=anthropic/claude-3.5-sonnet– specific LLM model
Final results are logged in outputs/YYYY-MM-DD/HH-MM-SS/, all program data is stored in the Redis DB.
Example execution:
# Basic run
python run.py problem.name=heilbron
# With experiment selection
python run.py problem.name=heilbron experiment=multi_island_complexity
# With parameter overrides
python run.py problem.name=heilbron max_generations=50 temperature=0.8 redis.db=5More details: See the "Customization" and "Configuration" sections in README.md for all available parameters and experiments
. Results Analysis
Results analysis — a set of tools for exporting and visualizing evolution data. Located in the tools/ directory. redis2pd.py extracts programs from Redis into CSV format with metrics and metadata for analysis in pandas/Excel; comparison.py generates a plot of the target metric evolution process for one or more experiments in PNG format. They work directly with the Redis DB and require matching redis.db and problem.name parameters with those used in the evolution run(s).
- Export to CSV (
redis2pd.py): Extracts all program data from Redis, including the target metric, additional metrics, generation numbers, and metadata. Outputs a flat CSV file with one row per evaluated program, ready for analysis in pandas, Excel, or statistical tools. Must match theredis.dband task name (prefix) from your evolution run. - Run comparison (
comparison.py): Generates a plot comparing the target metric evolution across one or more experiments in PNG format. Shows moving averages and standard deviations to illustrate convergence trends. Useful for comparing different experiment configurations (base vs multi-island), LLM models, or hyperparameters.
Both tools read directly from Redis, so ensure Redis is running and the specified database contains your evolution results.
Example usage:
- Export to CSV format
# Export a single run (matches redis.db and problem.name from run.py)
python tools/redis2pd.py \
--redis-db 0 \
--redis-prefix "heilbron" \
--output-file results/results.csv- Visualization of target metric evolution for one or multiple runs (PNG)
# Run several experiments with different DB numbers
python run.py problem.name=test redis.db=10 experiment=base
python run.py problem.name=test redis.db=11 experiment=multi_island_complexity
python run.py problem.name=test redis.db=12 experiment=full_featured
# Compare with plots
python tools/comparison.py \
--annotate-frontier \
--run "test@10:Baseline" \
--run "test@11:Multi_Island" \
--run "test@12:Full_Featured" \
--smooth-window 10 \
--iteration-rolling-window 10 \
--output-folder results/comparisonMore details: See tools/README.md for full documentation on analysis and visualization tools
Evolution of prompts for language models
In the new version, the GigaEvo Platform received a feature for automatic prompt enhancement for working with language models.
Creating an experiment with prompt evolution
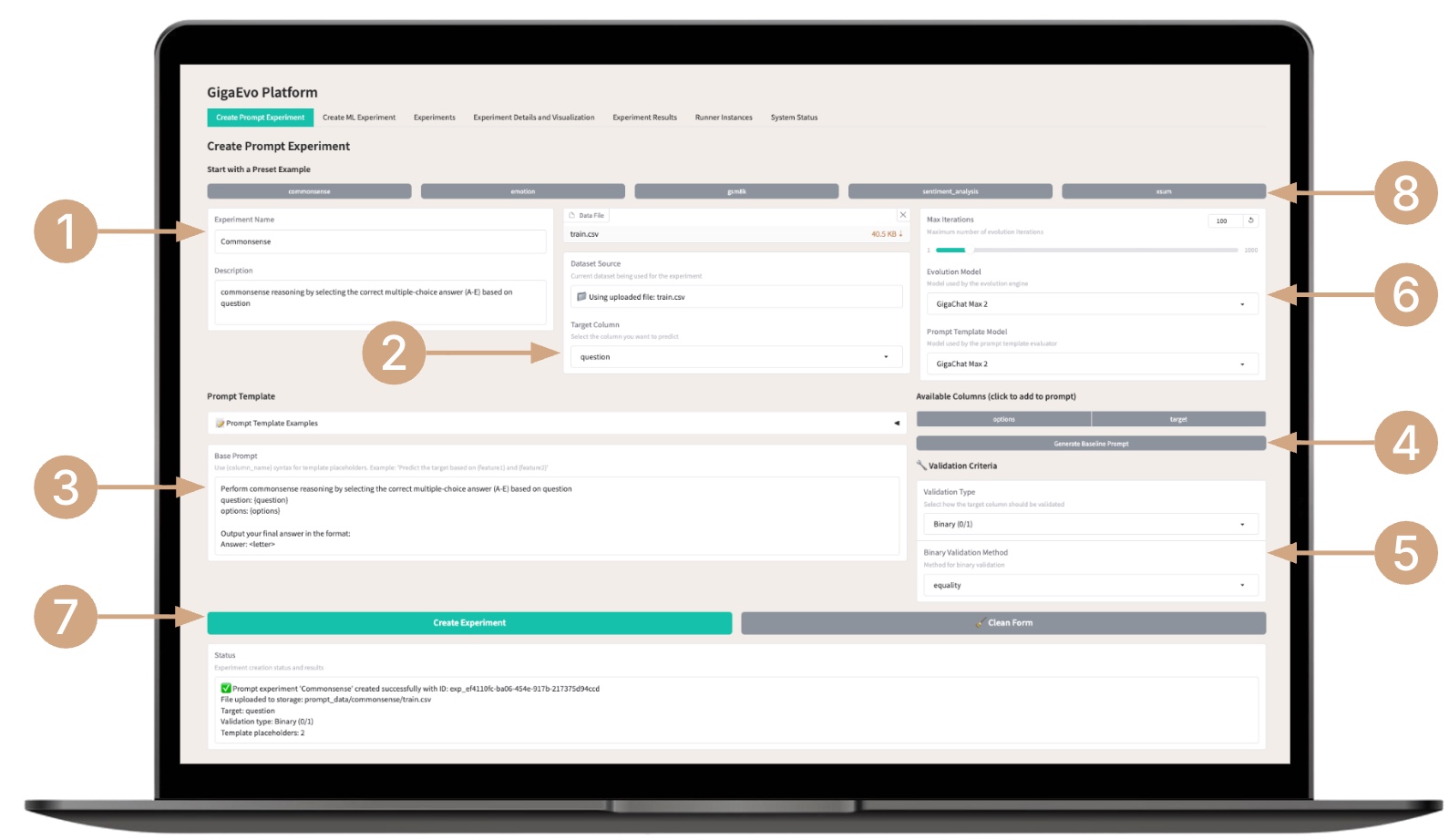
- Enter the title and a short description of the experiment’s goal.
- Upload a dataset and specify target metrics (target column).
- Write the base prompt for evolution (use data placeholders).
Your task: [describe what you want the LLM to do]
Context: {column1}, {column2}, {column3}
Question: What is the *target*?
Answer: [expected answer]
You are a sentiment analyst. Classify the review as positive or negative.
Review text: {review_text}
Rating: {rating}
Category: {product_category}
Answer : [expected answer]
- You can automatically generate the base prompt for evolution from a template or automatically add columns from the uploaded document.
- Select the type of validation (binary, continuous) and the evaluation metric relevant to your task:
Binary (Binary (0/1)) - for tasks with an exact answer
- Equality - exact match with the reference
- Substring - matching pairs of words
- RegExp - extract the answer using a given pattern
Continuous (Continuous (0..1)) - for text generation tasks
- ROUGE-1 - match of individual words
- ROUGE-2 - reference contained in the answer
- ROUGE-L - longest common subsequence
- BERTScore - semantic similarity
- BLEU - translation quality
- Configure additional parameters — number of generations, type of LLM model for evolution, and type of LLM model for validation.
- Create the experiment by clicking Create Experiment — the system will automatically assign it an identifier.
- You can also choose a template experiment from the list — the system will fill in the required fields and load the data; you’ll just need to run it.
The method works for any LLM-oriented tasks with verifiable results, for example:
- Text classification – adjusting prompts for sorting documents by category.
- Information extraction – improving prompts for identifying key data from text.
- Multi-step agent stages – optimizing prompts for complex systems with several working stages.
Using the results
- Load the experiment, start the evolution process, and wait for the best solution based on metrics.
- Export the experiment results — the platform will automatically select the prompt that provides the highest quality for your task.
- Use the resulting prompt to solve your target problem with your LLM.
Planned features
In upcoming versions, the following functionality will be added:
- Structured output formats (JSON, XML)
- Evaluation by another language model (LLM-as-a-judge)
- Custom metrics in Python
The feature is under active development.
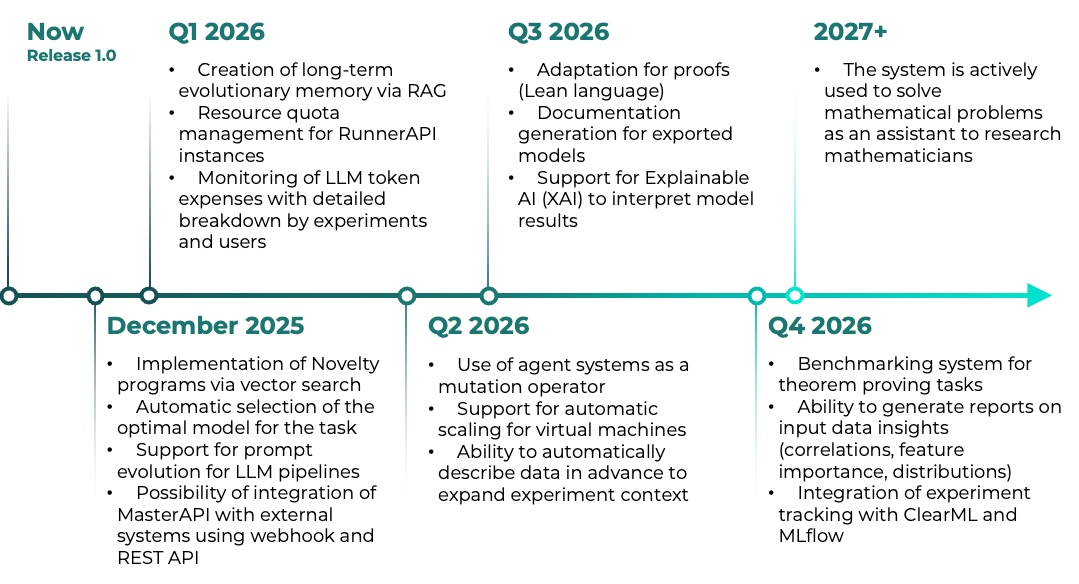
Check the project roadmap and stay tuned for updates!
Team

Aleksei Aleksandrovich Bokov
Research Engineer
AIRI

Olga Alekseevna Volkova
Junior Research Scientist
AIRI

Nikita Olegovich Glazkov
Junior Research Scientist
AIRI

Andrey Vladimirovich Galichin
Junior Research Scientist
AIRI

Valentin Andreyevich Khrulkov
Lead Research Scientist, Head of the "Generative Design" Group
AIRI

Igor Evgenyevich Trambovetsky
Systems Analyst
AIRI

Yaroslav Radionovich Bespalov
Research Scientist, Head of the "Multimodal AI Architectures" Group
AIRI

Bashkirov Denis Aleksandrovich
Sber, Risk block